|
CHROMOSOMES AND CHROMOCENTERS
IN THE GENUS Artemia
Trabajo recibido el 15 de febrero de 1989 y aceptado
para su publicación el 15 de agosto de 1989.
F. A. Abreu-Grobois.
Estación Mazatlán, Instituto
de Ciencias del Mar y Limnología, Universidad Nacional Autónoma
de México. A.P. 811, Mazatlán, Sinaloa. 82000
México.
J. A. Beardmore.
Department of Genetics, University College of
Swansea, Singleton Park, Swansea SA2 8PP, Great Britain. Contribución
No. 660 del Instituto de Ciencias del Mar y Limnología,
UNAM.
Se contó el
número de cromosomas y de cromocentros en preparaciones de
células somáticas de nauplios provenientes de un muestreo mundial
abarcando 23 poblaciones gonocorísticas (de cinco especies) Y 20
partenogenéticas del géneroArtemia. En las
especies gonocorísticas el número cromosómico diploide fue
de 42, con excepción deA. persimilis en la cual fue
de 44. En las dos especies gonocorísticas no estudiadas
citològicamente hasta ahora,A. monica y A. urmiana,
se encontraron también 42 cromosomas. Los niveles de ploidía
observados en material partenogenético fueron 2n, 3n, 4n, y 5n, con
algunos casos de heteroploidía en polipoides. No se encontraron
cromocentros en preparaciones de formas partenogenéticas, ni
deA. salina, ni de A. urmiana. Se
observaron cromocentros en células de todas las muestras de poblaciones
americanas. El promedio mínimo se encontró en A.
persimilis (3.03), mientras que el de A.
monicacayó dentro del rango observado para A.
franciscana(16.7-17.7). En las muestras de estas dos últimas
especies, se distinguen tres grupos con base en el número promedio de
cromocentros: (a) el Caribe (4 pobs.); (b) Yavaros, Sonora, México; y
(c) las restantes (13 pobs.). Se analiza el significado de los distintos
niveles de cromocentros en la evolución de las poblaciones y especies
del género.
Chromosome and
chromocenters were counted in naupliar somatic cell preparations extracted from
a world-wide sampling of 23 gonochoristic (five species) and 20 parthenogenetic
populations belonging to the genus Artemia. Diploid
chromosome numbers in populations of gonochoristic species including two
previously unstudied cytologically (A. monica and
A. urmiana, were all 42, with the exception of A.
persimilis which was 44. Ploidies observed in parthenogenetic
material included 2n, 3n, 4n, and 5n, with some cases of heteroploidy in the
polyploids. Chromocenters were absent in preparations of cells from
parthenogenetic froms, from A. salina and from A.
urmiana. Chromocenters were found in cell preparations from all New
World samples. Lowest numbers (mean= 3.03) were found in A.
persimilis. Mean chromocenter numbers in A. monica
(16.7) were within the range observed forA. franciscana
samples (mean range 16.7-17.7). Within last two species three population
clusters could be distinguished: (a) Caribbean (4 pops.); (b) Yavaros, Sonora,
Mexico (1 pop.);-and (c) remaining franciscana and monica samples (13 pops.) on
the basis of chromocenter numbers. The evolutionary significance of the
chromocenter number clusters is discussed.
The anostracan branchiopods of the genus Artemia are widely distributed throughout the world where natural or man-made salterns are found. Their capacíty for dispersal depends on the accidental adherence of viable cysts to migrating birds feeding in their habitants (MacDonald, 1980) or on the deliberate spread by man (Persoone and Sorgeloos, 1980). This genus includes four gonochoristic species: A. franciscana Kellog, 1906 (found in habitats in the Americas except Argentina), A. persimilis Piccinelli and Prosdocimi, 1968 (found naturally only in Argentinan habitats), A. salina(Linnaeus, 1758) Leach, 1819 (in European and North African slaterns) andA. urmiana Günther, 1900 (in Lake Urmia, Iran). A fifth gonochoristics species, A. monica Verrill, 1869 is found only in alkaline Mono Lake, California, USA where it is ecologically isolated (Bowen et al., 1985). Natural populations of parthenogenetic forms of various ploidies, probably evolving from a common ancestor to the A. urmianaline (Beardmore and AbreuGrobois, 1983) frequently coexist in Mediterranean and Middle Eastern salterns. At times and in some sites A. salina is present as well (cf. Artom, 1921; Gross, 1932; Stefani, 1967; Browne and MacDonald, 1982). The diploid chromosome numbers of A. salina and A. persimilis, have been determined by Piccinelli and Prosdocimi (1968) to be 42 and 44 respectively. Tetraploidy inA. franciscana from Great Salt Lake has been reported (Barigozzi and Tost, 1959) but later studies (Iwasaki, 1969; Barigozzi and Baratelli-Zambruni, 1982) found the population to be diploid. Among parthenogenetic forms a range of polyploid numbers (3n, 4n, 5n) have been found as well as the more common diploid condition (Barigozzi, 1974). Cytological observations of resting nuclei have also revealed the presence of chromocenters exclusively inA. franciscana (Bargigozzi and Baratelli- Zambruni, 1981) The present work aims to extend the number ofArtemia populations surveyed for both chromosome and chromocenter numbers and include two cytologically unstudied biparental species: A. monica and A. urmania. MATERIALS AND METHODSArtemia samples were collected and transported as cysts from localities listed in Tables I and 11. For the cytological preparations a modification of the methods of Barigozzi and Baratelli-Zambruni (1981) and Barigozzi et al. (1984) was employed, incorporating some elements described by Kligerman and Bloom (1977). Briefly, newly hatched nauplii (see Sorgeloos et al., 1986 for hatching methods) were subjected to hypotonic shock in distilled water for 1 hr. at 30°C. Nauplii where then fixed in 1:1 methanol -glacial acetic acid at room temperature for 3 mins after which the liquid was removed and replaced with 60% acetic acid at 45ºC. Individual nauplii, which during the subsequent 20-30 seconds swell and break up, were transferred to separate positions on one or more slides with enough acetic acid to form 5-10 mm circles. Cell preparations were dried at 45ºC and stained on horizontal racks with freshly diluted and filtered Giemsa stain (Gurr improved R66 solution, 1:3 stain to 0.1M phosphate buffer pH 6.8) for 7-10 mins. Stained slides were mounted with DePeX mounting medium after rinsing with distilled water and air-drying. Chromosome counts (at 1000x magnifícation) were made from at least 8 nauplii, from each population. Chromocenter observations were made on 4 nauplii, counting 10 nuclei from each. Populations designated "parthenogenetic" in this work were those which were either verified to reproduce asexually in cultures or of having genotypes assignable to parthenogenetic forms observable in enzyme electrophoresis assays (Abreu-Grobois, 1983). A set of samples from two habitats (San Francisco Bay and Great Salt Lake) were obtained 5 and 10 years apart, respectively, and their results are reported separately in this study. Identification of sexual species followed morphological traits enlisted in Beardomore and Abreu-Gorbois (1983). RESULTS
CHROMOSOME NUMBERResults of the chromosome counts are entered in Tables III and IV. Among the gonochoristic material, samples from A. franciscana (figures 1 and 2) andA. salina populations were all found to contain diploid chromosome numbers (2n=42, Table III) in agreement with earlier reports. Only diploid Nauplii (Table III) were found in the BAR population sample although tetraploid forms have also been reported here under natural conditions (Amat Domenech, 1980); these were assigned to A. salina on the basis of their characteristic electrophoretic pattern observed in separate investigations (Abreu-Gobois, 1983). The remaining two gonochoristic speciesA. monica and A. urmiana were both also diploid. The only case of heteroploidy found among biparental populations was in GSL material with one metaphase count of 43 chromosomes. All ploidy levels in parthenogenetic populations (Table 4; figures 3,4) reported previously (see Barigozzi, 1974 for review) were observed. Out of the 21 localities where unisexual forms were collected, 13 contained single ploidies (61% of the total), while the remaining 8 contained mixtures of di- and tetraploids (24%, 5 in all), diand pentaploids (10%, 2 in total), or tri-tetra- and pentaploids (5%, 1 population). Among sites where parthenogeneticArtemia was found, diploid forms were by far the most frequent, observed in 57% of the populations assayed. Heteroploidy among unisexualArtemia was observed particularly in anisopolyploids: EIL, IZM, TIEN, and TUT (Table 4.) 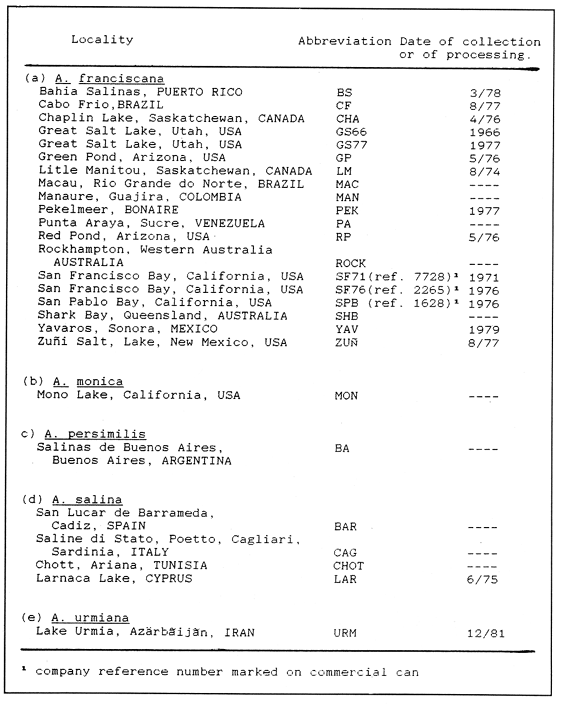 TABLE 1 BISEXUAL Artemia POPULATIONS SAMPLED 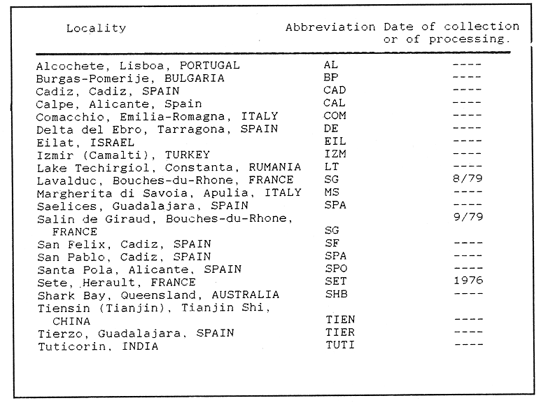 TABLE 2 PARTHENOGENETIC Artemia POPULATIONS SAMPLED CHROMOCENTERSIn agreement with Barigozzi and Baratelli Zambruni (1981), chromocenters in resting cells were found in A. franciscana material (Table 3; figures 2 and 5). However, they were also present in A. persimilis (figure 7) and in A. monica which had not been reported previously. No such structures were found in A. salina, in A. urmania , nor in any of the parthenogenetic populations (Table 4; figure 6 for example). Among the American samples, a range of chromocenter numbers were found (Table 3). A. persimilis contained the lowest number (mean = 3.03, figure 7) among the New World species. However, YAV (figure 8), an A. franciscana population also had very low number of chromocenters, being thus similar in this respect to A. persimilis. The A. franciscanaand A. monica population may be divided into three clusters on the basis of their chromocenter numbers: cluster I, containing all the North American populations (CHA, GS66, GS77, GP, LM, RP, SF76, SPB, MON) or transplants from San Francisco genomes (ROCK, MAC, SHB), but excluding YAV, with an overall mean of 17.08 and a range of 11-24 chromocerters; cluster II, the Caribbean populations (BS, MAN, BON, PA) with a mean of 17.22 and a range of 4-14; and, cluster III, the YAV population, with a mean of 3.90 and a range of 2-6. Chromocenters in cluster I not only appeared more abundant but also considerably large in size in interphase preparations (compare figures 2 and 5 ). A very small ANOVA F value for the North American population chromocenter counts (Table 5) is indicative of large variation within each sampling. However, no significant heterogeneity was observed between populations within each group. The distribution of values for the cluster I populations, shows a significant skewness to the right (g1 = 0.35, ts = 3.35, P .001) but no significant kurtosis (g2 = 0.17, ts = 1.6, n.s.) which suggests that the actual number are higher than the mean reported here but some overlapping of chromocenters inevitably occurs in the heterochtomatic masses to be concentrated on the ends of some of the chromosomes. 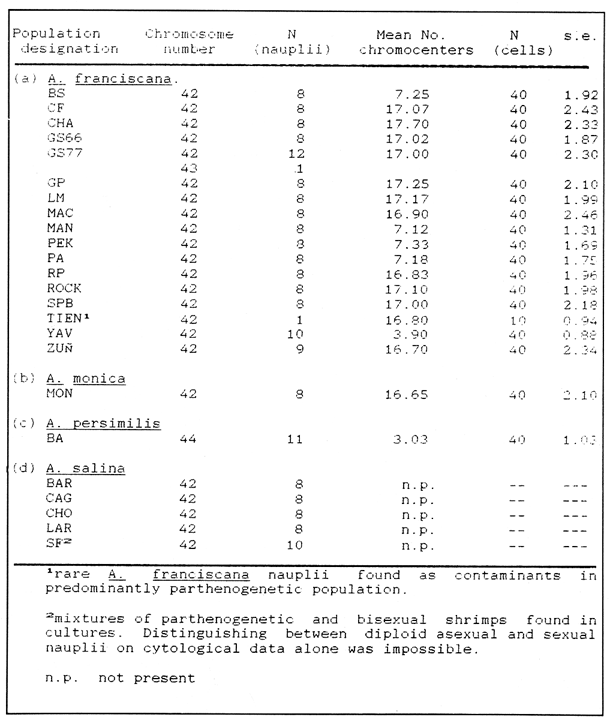 TABLE 3 CHROMOSOMES AND CHROMOCENTERS IN BISEXUAL Artemia 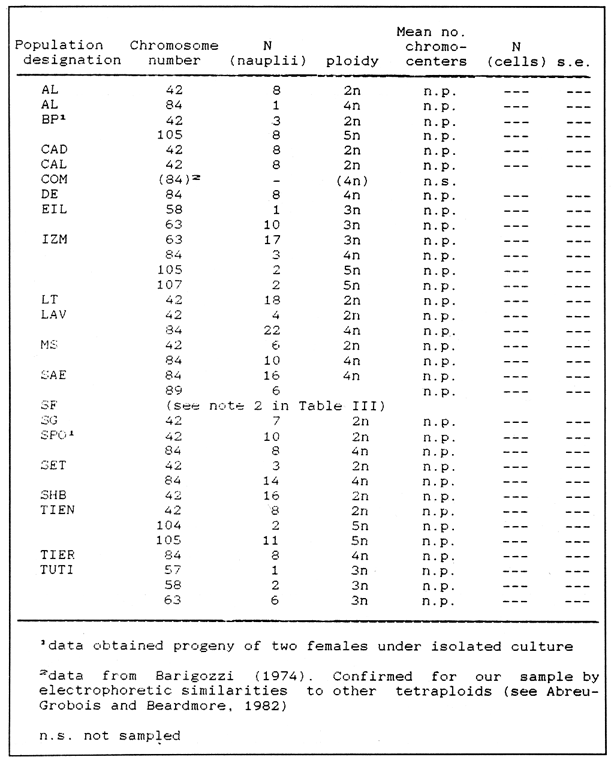 TABLE 4 CHROMOSOMES AND CHROMOCENTERS IN PARTHENOGENETIC Artemia 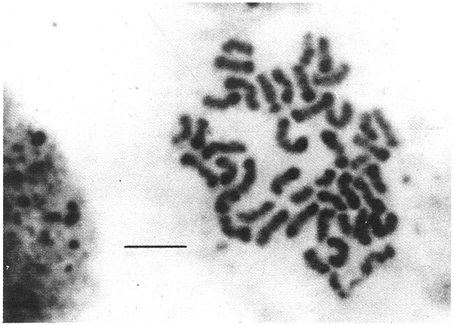 Figura 1. Prophase plate from A. franciscana of GS77, Bar represents 5 micron. 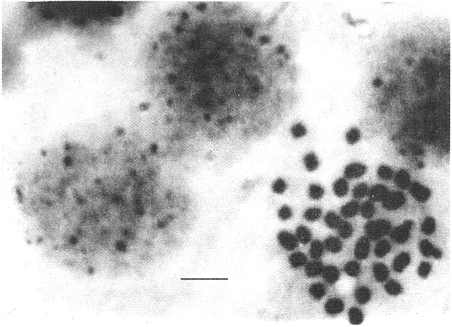 Figura 2. Cell preparation of material from A. franciscana of CUR (2n-42). 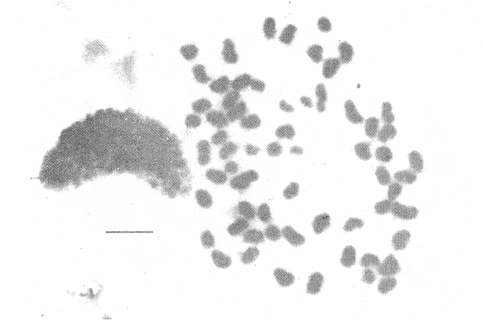 Figura 3. Metaphase plate from Artemia sp. of EIL (3n=63). 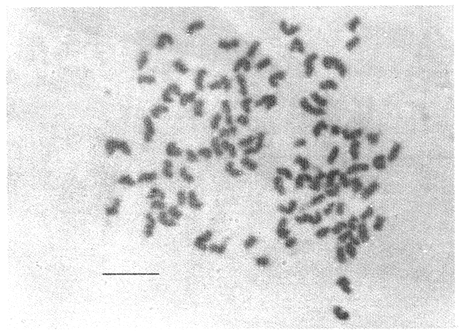 Figura 4. Metaphase plate from Artemia sp. of TIEN (5n=105). 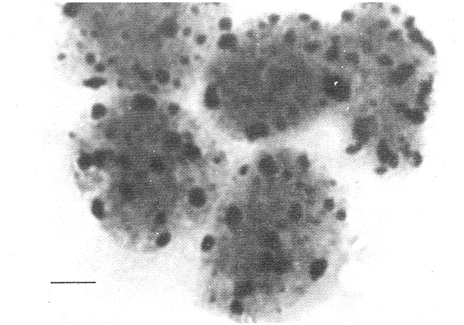 Figura 5. Interphase nuclei from GS66 (A. franciscana). 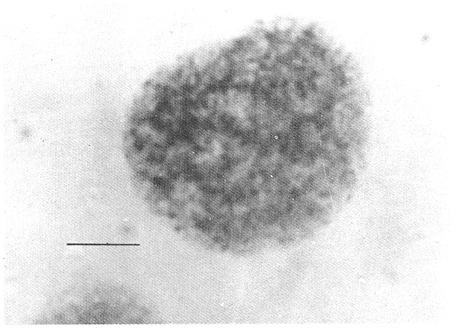 Figura 6. Interphase nuclei of parthenogenetic Artemia from SG.. 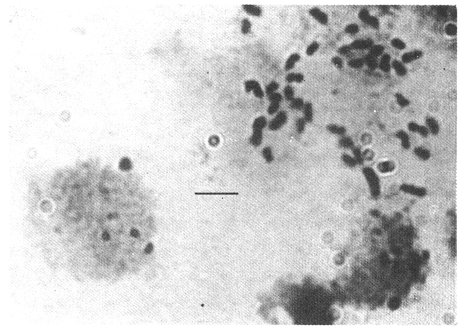 Figura 7. Chromocenters in interphase nuclei of A. persimilis from BA. 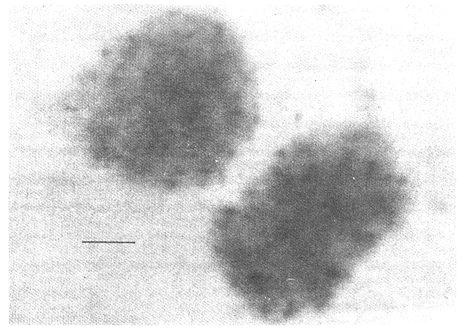 Figura 8. Chromocenters in preparation from Yavaros A. franciscana. 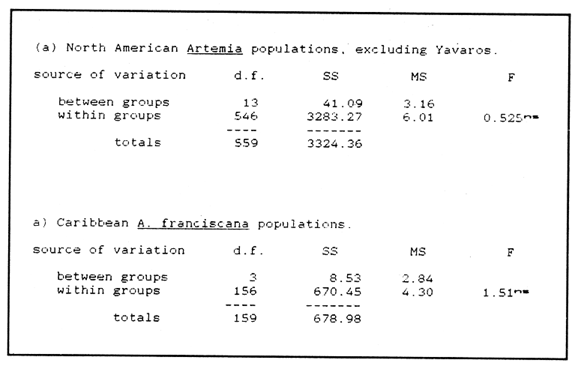 TABLE 5 ANOVA FOR CHROMOCENTER SURVEYS DISCUSSIONThe much broader geographic range of parthenogenetic populations in the Old World observed in the material studied probably reflects a greater capacity for colonizing newly opened environments that asexual Artemiaforms have, compared to their sexual relatives, at least, in that region of the world. This is in agreement with Browne and MacDonald (1982). Furthermore, being able to distinguish the range of the parthenogenetic forms by ploidy levels permits a similar obervation regarding the diploid asexualArtemiawhich are much more widely dispersed than their polyploid conterparts, suggesting that polyploidy per sedoes not confer this organism any additional capacity for colonizatíon and population spread. The presence of chromocenters in a restricted number of species makes their visualization useful for identifying species or strain mixtures in cyst samples reasonably quickly and cheaply. For example, the TIEN sample contained A. franciscana nauplii in low frequencies and these could be determined by chromocenter analysis (Table 3). Most importantly, the clear differences in numbers of chromocenters found between and within species also have evolutionary significance. The differences observed between populations are characters that distinguish between the Old and New World gonochorisitc species and between the Caribbean and the North American (A. franciscana and A. monica) populations. The grouping based on this cytological feature closely parallels dendrograms drawn from electrophoretic studies (Abreu-Grobois and Beardmore, 1982). The distinction between the YAV, population and the remainingA. franciscana A. monica populations found in this cytological survey is not so clear in electrophoretic: analyses, although low heterozygosity values for this population compared to the rest of the species (Abreu-Gobois, 1983) is indicative of having undergone recent bottlenecks. Events such as these could also explain the marked cytological differences observed for this population. The nature of chromocenters can be deduced from the use of by quinachrine (Barigozzi and Baratelli Zambruni, 1981) and Giemsa dyes (Barigozzi et al., 1984; this study) which commonly stains preferentially areas of DNA rich in A-T residues. More detailed research with material extracted from A. franciscana collected from the same locality in San Francisco Bay as that used for the present study has shown the chromocenters visualized with these dyes to constitute heterochromatic blocks in the chromosomes, rich in repetitive Alu I 110-bp fragments (Barigozzi et al., 1984). It is posible to speculate on the evolutionary derivation of the chromocenter numbers. If lack of chromocenters in the Old World forms is the primitive state, then amplification and interchromosomal spread of the same sequence(s) from an original common ancestor through a mechanism such as that submitted by Dover (1982) must have occurred in the New World species after their separation from a common ancestral lineage. For the opposite to be true, presence of chromocenters being the primitive state, a mechanism of repeated deletions would be necessary to explain the condition of the Old World fórms. Present data cannot be used in choosing between these hypothetical evolutionary schemes involving the split between New World and Old World Artemia lines. On the other hand, while it is not certain whether the stucture of the repetitive DNA in all of the New World Artemia populations is equivalent and the same as that found by Barigozzi et al. (1984) in SF samples, the fact that the pattern of different chromocenter numbers in populations within and between closely related species in the New World closely parallels their overall genetic distances (see Abreu-Grobois and Beardmore, 1982) suggests either their causative involvement in the mechanism(s) of differentiation and speciation or, if the chromocenters are neutral to these processes, they do trace changes at the intragenomic level which the taxa has experienced. For example, if the heterochromatic blocks inA. persimilis are a subset of these in the Caribbean populations of A. franciscana and the latter, in turn, are a subset of these in populations of the same species in North America, then duplication and amplification of a common ancestral repetitive sequence could have been involved. The differences in the quantities of the chromocenters would imply different rates of spread and/or amplification in the differentArtemia groups. Alternatively, if the sequences of the chromocenters in the different clusters of populations turn out to be different, then population-specific amplification of the same or distinct sequence repeats would be involved. Studies in other organisms have pointed out the striking rapidity in which repeated sequences can spread to individuals of a single species (Brown et al., 1972). Amplification of particular repeated sequences can cause vital alterations in the genetics ol organisms: repeated DNA's have effects on recombination (John, 1981) and on other cytogenetic properties causing large-scale effects on gene expression (Peacock et al., 1982) or even possibily providing a driving force for speciation to ocurr (Dover, 1982). Further studies are necessary to ascertain the role repeated sequences have played in the differentiation of theArtemia populations. As may be expected, no differences in chromocenter numbers were found between samples of nauplii from habitats taken up to 11 years apart (GS66 and GS77). However, the time sequences involved in the establishment of the differences observed may be studied sinceArtemia cysts are known to withstand burial in salt sediments for thousands of years. Conclusiones1. Diploid chromosome number in gonochoristicArtemia species (including A. urmiana andA. monica) both previously unstudied cytologically) was found to be 42, with the exception of A. persimilis where it was 44. 2. Diploid parthenogenetic forms are more widely distributed geographically than either polyploid asexuals or A. salina, both commonly found within the range of the latter. 3. Among the Old WorldArtemia, the greater geographic range of diploid parthenogens in Mediterranean Europe and Africa is suggestive to their greater colonizing capacity than either A. salina or polyploid parthenogens. 4. Chromocenters, structures found only in New World Artemiaare useful in identifying presence of American species in mixed samples of cysts. 5. Clustering ofA. franciscanaand A. monica) populations according to mean chromocenter numbers into three groups: Yavaros (Sonora, Mexico); Caribbean populations; and remaining populations closely parallels the genetic differentiation observed on this group utilizing other analytic methods and suggests strong intraspecific evolutionary divergence involving repetitive DNA sequences. AgradecimientosThe present research was partly funded by a scholarship from CONACYT, Mexico awarded to one of the authors (FAAG). We would like to thank the following people and institutions who kindly contributed cysts: Francisco Amat- Domenech,Artemia Reference Center, Sarane Bowen, Nick Collins, Gayle Dana, Alberto Ramirez-Flores, Ali Valamanesch, Graham Wallis, Robert D. Ward. The pertinent remarks of an anonymous reviewer greatly improved this report. LITERATURAABREU-GROBOIS, F. A. Ph. D. Thesis Population genetics of Artemia.Univ. Coll. Swansea, Great Britain.1983438 p. ABREU-GROBOIS, F.A. and J.A. BEARDMORE Genetic differentiation and speciation in the brine shrimp Artemia. C. Barigozzi Mechanisins of Speciation. Alan R. Liss, Inc. New York1982p. 345-376, 546 p. AMAT DOMENECH, F. The Brine Shrimp Artemia.Differentiation in Artemia strains from Spain. p. 19-39. G. Persoone, P. Sorgeloos, O. Roels, and E. Jaspers. Universa Press, WetterenBelgium1980345 p. Vol 1. ARTOM, C. Specie micropireniche e macropireniche del genre Artemia. Ric. Morfol1921137-1552 BARIGOZZI, C. Artemia: a survey of its significance in genetic problems. Evalut. Biol.1974221-2527 BARIGOZZI, C. and L. BARATELLI-ZAMBRUNI Presence and absence of chromocenters in populations of Artemia.Atti. Acad. Naz. Lince1981122-1251 BARIGOZZI, C. and L BARATELLI-ZAMBRUNI New data on chromosome number of the genus Arteinia. Atti. Acad. Naz. Lincei.1982139-14373 BARIGOZZI, C. and M. TOSINew data on tetraploidy of anphigonic A. salina Leach and on triploids resulting from crosses between tetraploids and diploids.Conv. Genet. Suppl. Ric. Sci.19593-629 BARIGOZZI, C., G. BADARACCO, P. PLEVANI, L. BARATELLI, S. PROFETA, E. GINELLI, and R. MENEVERI Heterochromatin in the genus Artemia. Chromosoma(Berl.)1984332-33790: BEARDMORE, J. A. and F. A. ABREU-GROBOIS Protein Polymorphism: Adaptive and Taxonomic Significance. Taxonomy and evolution in the brine shrimp Artemia. G.S. Oxford and D. Rollinson Systematics Association/Academic PressLondon1983p. 153-164, 405 p. BOWEN, S.T., E.A. FOGARINO, K.N. HITCHNER, G.L. DANA, V.H.S. CHOW, M.R. BUONCRISTIANI, J.R. CARLEcological isolation in Artemia: population differences in tolerance of anion concentrations.J. Crust. Biol.1985106-1295 BROWN, D.D., P.C. WENSINK, and E. JORDAN A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes.J. Mol. Biol.197257-7363 BROWNE, R.A. and G.H. MACDONALDBiogeography of the brine shrimp Artemia: distribution of parthenogenetic and sexual populations.J. Biogeogr.1982331-3389 DOVER, G. Mechanisins of Speciatioti. The role for the genome in the origin of species. C. Barigozzi Alan R. Liss, Inc.New York1982p. 435-460, 546 p. GOLDSCHMIDT, E.Fluctuation in chromosome number in Artemia salina.J. Morphol.195211-13491 GROSS, F. Untersuchungen u die Variabilitat bel Artemia salina.Naturwissenschaften1932962-96720 GUNTHER, R.T.Contributions to the natural history of Lake Urmi, N.W. Persia, and its neighborhood. J. Linnean Soc. Zool.1900345-45227 IWASAKI, T. Chromosome number of Artemia salina obtained in the Great Salt Lake. UtahJ. Genet.U.S.A. Japan.1969105-10644 JOHN, B. Chromosomes Today Heterochromatin variation in natural populations.1981128-1377 KELLOG, V. A new Artemia and its life conditions.Science1906594-59624 KLIGERMAN, A.D. and S.E. BLOOMRapid chromosome preparations from solid tissues of fishes. J. fish. res. Bd. Can.1977266-26934 LEACH, W. E.Entomostraca. Dictionarie des Sciences Naturelles.1819p. 524-543 Vol. 4. MACDONALD, G.H. The Brine Shrimp Artemia The use of Artemia cysts as food by the flamingo (Phoenicopterus ruber roscus) and the shelduck (Tadorna tadorna). G. Persoone, P. Sorgeloos, O. Roels, and E. Jaspers Universa PressWetteren Belgium.1980 p. 97-104. Vol. 3. PEACOCK, WJ., E.S. DENNIS, and W.L. GERLACH Mechanisms of Speciation. DNA sequence changes and speciation. C. Barigozzi Alan R. Liss, Inc. New York1982p. 123-142 546 p. PERSOONE, G. and P. SORGELOOSThe Brine Shrimp Artemia General aspects of the ecology and biogeography of Artemia G. Persoone, P. Sorgeloos, 0. Roels, and E. Jaspers Universa PressWetteren, Belgium1980p. 3-24. Vol. 3. PICCINELLI, M. and T. PROSDOCIMIDescrizione tassonomica delle due apecie Artemia salina L. e Artemia persimilis n. sp.Inst. Lomb. Sci. Lett.1968113-118102 SORGELOOS, P., P. LAVENS, P. LEGER, W. TACKAERT, and D. VERSICHELE Manual for the Culture and Use of Brine Shrimp Artemia in Aquaculture. State University of Ghent, Belgium.1986319p. STEFANI, R. La maturazione dell'uovo nell' Artemia salina di Sete.Riv. Biol.1967599-61560 VERRILI, A. E.Observations on phyllopod crustacea of the family Branchiopidae, with description of some new genera and species from America.Proc. Am. Assoc. Advan. Sci.1869230-247118
|

